Proteasomal protein degradation is crucial in maintaining cellular integrity and in regulating key cellular processes including cell cycle, proliferation and cell death. The 20S proteasome can function as an independent degradation machinery and tight regulation must exist to control its proteolytic capacity. Despite the major importance of 20S proteasome regulation only a small number of regulators have been identified to date. Here, we discovered a novel family of inhibitory protein - CCRs (Catalytic Core Regulators) that specifically regulate the 20S proteasome, and not the 26S counterpart, and increase its effectiveness in cancer treatment.
The protein degradation machinery plays a critical role in the maintenance of cellular homeostasis, preventing the accumulation of damaged or misfolded proteins and controlling the levels of regulatory proteins. Proteasomal degradation is mediated mainly by two proteasomal complexes: the 26S proteasome and the 20S proteasome which under oxidizing conditions is the major degradation machinery. While the presence of a regulatory mechanism to control this degradation process is highly likely, only two 20S proteasome regulators have been identified; NAD(P)H dehydrogenase [quinone] 1 (NQO1) and DJ-11.
Prof. Michal Sharon and her team discovered a novel family of Catalytic Core Regulators (CCRs)2 that controls 20S proteasome-mediated degradation. These could be used for cancer therapy and treatment for diseases associated with proteasome dysfunction.
The team performed a bioinformatic screen to identify other proteins with sequence and structural similarities to DJ-1 and NQO1. This led to the discovery of a family of 17 relatively small proteins of 20–30 kDa, named catalytic core regulators (CCRs), that oversee 20S proteasome activity. Of the 10 short-listed proteins that were identified and characterized, all were able to inhibit 20S proteasome degradation of known substrates, both in vitro and in cells. These protein regulators were able to specifically bind to the 20S proteasome, but not the 26S proteasome, and affect protein degradation. In addition, they demonstrate that the CCRs organize into a feed-forward regulatory circuit3 involving the master regulator of the oxidative stress response, Nrf2. Certain CCRs influence the stability of Nrf2, which subsequently upregulates the expression of other CCRs, leading to an overall dampening of 20S proteasome-mediated degradation of unfolded protein substrates within the cell.
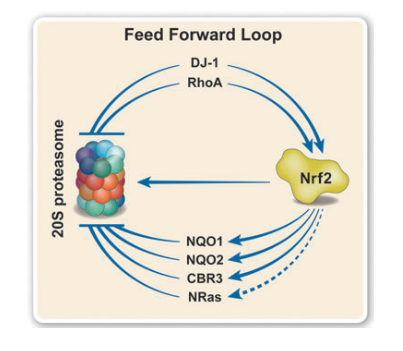
Figure 1: CCRs organize into a feed-forward regulatory loop with Nrf2 and the 20S proteasome.
Prof. Sharon and her team found that CCRs bind the proteasome through an internal b-strand that is exposed upon interaction with the 20S proteasome. Binding by the 20S proteasome is mediated by the b-strand rather than the a-ring, and specifically by the PSMB4 subunit. Moreover, they discovered that the interaction formed by PSMB4 and the CCR leads to allosteric inhibition of the proteasome’s three enzymatic activities. By dissecting the structural properties that are required for CCR-like function, the group could recapitulate the CCR activity using a designed artificial protein that is half the size of natural CCRs. This mean of 20S proteasome-specific attenuation opens avenues for decoupling the 20S and 26S proteasome degradation pathways for selective 20S proteasome-inhibitors for therapeutic applications.
References:
1. Moscovitz, O., Ben-Nissan, G., Fainer, I. et al. (2015). The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat Commun 6, 6609 https://doi.org/10.1038/ncomms7609
2. Olshina, M. A., Arkind, G., Kumar Deshmukh, F., Fainer, I., Taranavsky, M., Hayat, D., Ben-Dor S., Ben-Nissan G.,Sharon, M. (2020). Regulation of the 20S proteasome by a novel family of inhibitory proteins. Antioxidants & redox signaling, 32(9), 636-655. https://doi.org/10.1089/ars.2019.7816
3. Mangan S, Alon U. (2003). Structure and function of the feedforward loop network motif. Proceedings of the National Academy of Sciences, 100(21), 11980-11985. https://doi.org/10.1073/pnas.2133841100

