Inflammatory bowel disease (IBD) patients suffer from chronic inflammation of the gastrointestinal (GI) tract. The disease course varies, and patients often experience relapses and remission, despite the availability of several medications types. Currently, the disease stage is only assessed by histological analysis of colonoscopy obtained biopsies. Therefore, there is a critical need for non-invasive diagnostic and prognostic markers for IBD.
Prof. Itzkovitz and his team show that the feces transcriptome of host shed cells is a diagnostic biomarker for GI histological inflammation and could possibly be used as a prognostic tool to predict patients' responses to different treatments.
IBD (Crohn’s and ulcerative colitis) is characterized by chronic GI inflammation. IBD prevalence in Israel and worldwide is rising. Several types of medications exist, including biological therapies that have greatly improved patients’ condition. However, many patients are either primary non-responders or experience loss of response over time, resulting in disease flares. Initial diagnosis and monitoring of the disease are often based on assessing histological inflammation of the GI, using biopsies obtained upon colonoscopy. However, colonoscopies are cumbersome, expensive, require unpleasant preparation, and are also associated with potential complications of perforation/sedation. Existing non-invasive prognostic tools, including stool proteomics and micriobiome characterization are limited in their diagnostic power. Therefore, there is a critical need for non-invasive markers for the state of GI inflammation that could be used to predict patient prognosis or response to therapies.
Prof. Shalev Itzkovitz and his team showed that transcriptomics of patients’ fecal samples are correlated with the inflammatory state of the intestine and could be used as a non-invasive diagnostic and prognostic tool for IBD.
Prof. Itzkovitz and his team argued that RNAseq measurements of the host transcripts, originating in cells that are constantly shed into the lumen, could be a proxy for inflammation status. They performed RNA sequencing of biopsies and fecal-wash samples from IBD patients and controls undergoing lower endoscopy. They showed that RNAseq of the host transcripts in fecal washes contained different information compared to RNAseq of biopsies. Importantly, the Fecal wash host transcriptome was significantly more informative than the biopsy transcriptome in identifying histological inflammation and classifying patient disease status (Figure 1). This increased power of fecal transcriptomics stems from the fact that the feces represent cells that are shed throughout the gastrointestinal tract. Based on these results, the researchers identified specific gene signatures in fecal washes samples from patients with and without histological inflammation, which could be used as a diagnostic tool for classifying patients’ disease status. The team also analyzed fecal wash proteomics using Mass Spectrometry and showed that fecal wash transcriptomics carries distinct information from fecal proteomics.
Therefore, the transcriptome of patients’ feces is a powerful non-invasive biomarker that reflects histological inflammation. In addition to being a diagnostic tool, identifying correlations between feces-transcriptomic signatures and response to specific therapies could potentially provide a new, personalized prognostic tool.
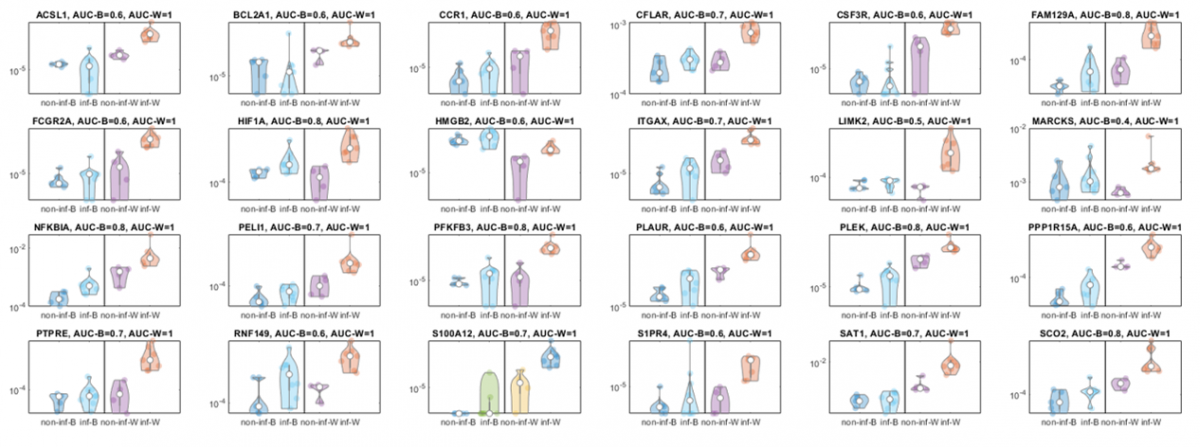
Figure 1- Distributions of gene expression levels in biopsies and in fecal washes of ulcerative colitis (UC) patients with histological inflammation versus UC patients with histological healing. Each panel shows a host gene, Y axis is mRNA expression as a fraction of all sample reads, white circle denotes the median, left parts of each plot show the distributions of gene expression in the non-inflamed biopsies (non-inf-B) and the inflamed biopsies (inf-B), right part shows the gene expression levels in the non-inflamed washes (non-inf-W) vs. the inflamed washes (inf-W). AUC – Area under the curve of ROC analysis.Fecal wash samples, yet not biopsies, harbor dozens of genes with AUC=1, enabling error-free classification of histological inflammation state.
- A non-invasive diagnostic tool for IBD
- A personalized method of selecting the optimatl treatment for IBD patients
- Patient-friendly – a fecal wash sample, potentially obtained using a home stool kit for patients’ use
Prof. Itzkovitz and his team demonstrated that feces transcriptome correlates with histological inflammation. They identified specific genes that are differentially expressed in fecal washes of IBD patients reaching histological healing and those with histological inflammation. Future work will further validate the results and identify specific genetic patterns in pre-therapy fecal washes which correlate with response to different therapies (patient-specific tailoring of biological therapy). Finally, the fecal wash assay will be adjusted into a home stool kit for patients’ use.

