Antibiotic-resistant bacteria such as Staphylococcus aureus (MRSA) are a growing threat, causing severe infections that are increasingly untreatable by existing drugs. This technology provides a high-resolution crystal structure of the large ribosomal subunit (50S) from pathogenic S. aureus, enabling structure-based discovery of new antibiotic classes. Derived from a clinically relevant strain, the structure reveals novel, species-specific binding sites that can be exploited for precision antibiotic development.
- Structure-guided design of novel antibiotics targeting S. aureus and MRSA
- Computational screening and virtual docking of compound libraries
- Rational optimization of existing antibiotic scaffolds
- Structural biology resource for elucidating bacterial ribosomal mechanisms
- First high-resolution ribosome structure from a pathogenic S. aureus strain
- Antibiotic-soaked crystal structures reveal drug–ribosome interaction sites
- Species-specific ribosomal differences provide unique antibacterial target
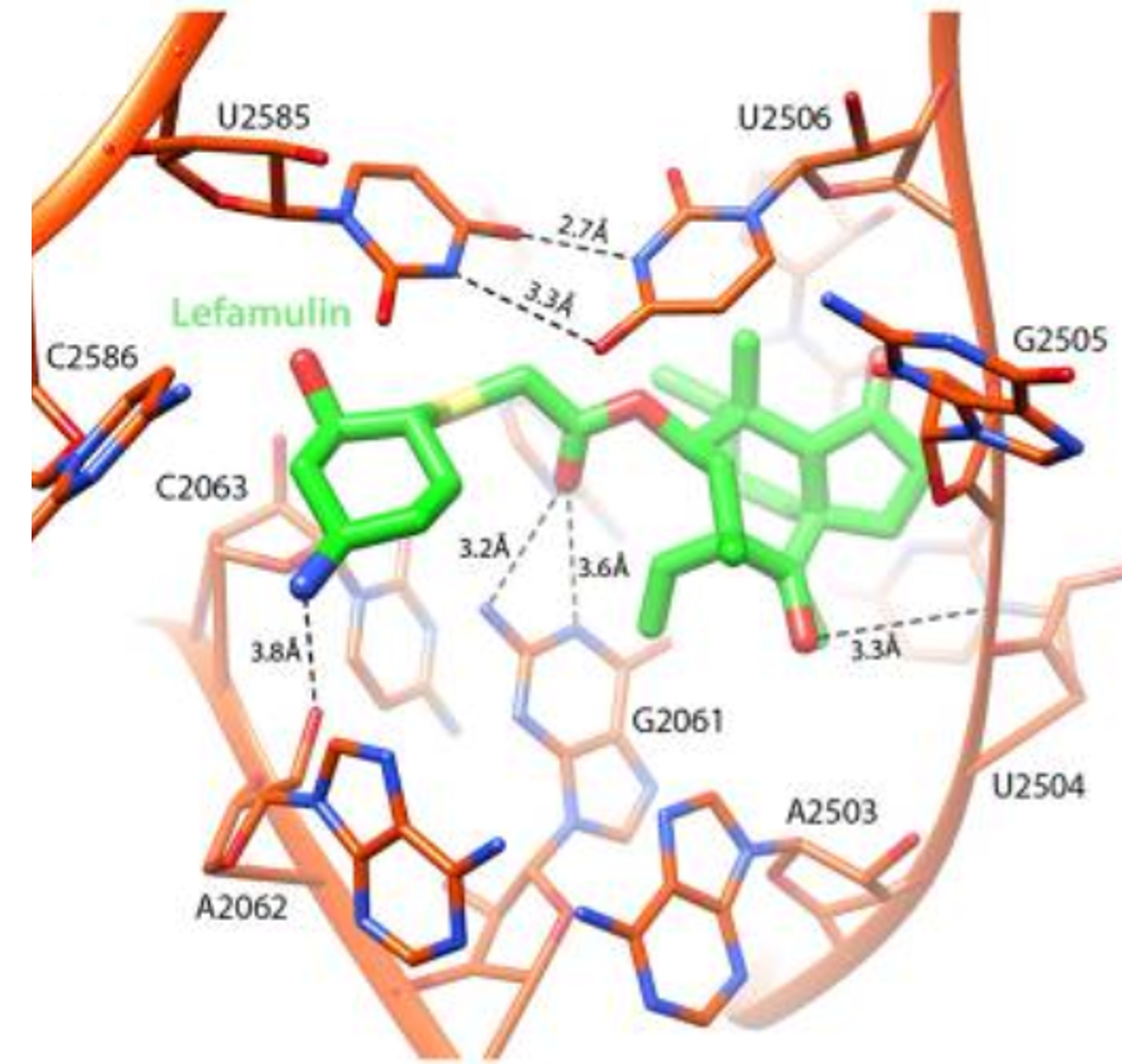
Zoom into the lefamulin (green) pocket in 23S rRNA (orange), showing U2585–U2506 interactions that stabilize binding and elucidate the antibiotic’s mechanism of action.
The 50S ribosomal subunit was crystallized from a pathogenic S. aureus strain, and several antibiotic-complex structures were resolved at high resolution, defining new binding pockets for rational drug design. The platform supports computational and experimental screening of next-generation antibiotic candidates.


