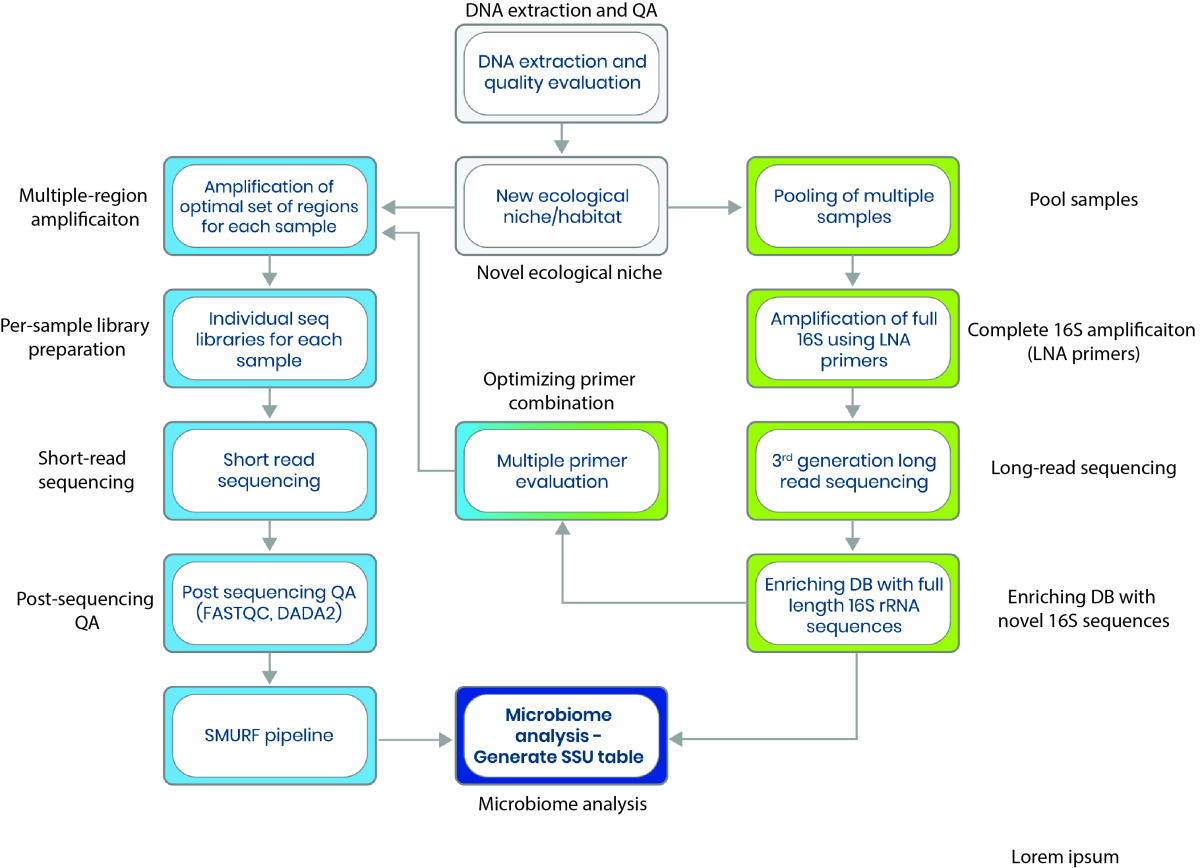
Microbial profiling methods based on 16S rRNA sequencing are limited by primer bias, short-read coverage, and databases dominated by human-associated microbes. These limitations reduce phylogenetic resolution and hinder accurate species detection in underexplored environments. CoSMIC is a hybrid profiling approach that combines long-read sequencing with short-read analysis over multiple 16S regions, enabling precise, cost-effective identification of microbial communities. It overcomes traditional biases and offers robust profiling even in low-biomass or fragmented DNA samples.
- Microbiome mapping in agricultural soils for precision farming (e.g., irrigation and pesticide optimization)
- Development of microbial inoculants to enhance seed, soil, or antimicrobial traits
- Surveillance of bacteria-prone environments (e.g., aquaculture, hospitals, food processing)
- Microbial monitoring in water treatment and quality assessment.
- Improved PCR-quantification fidelity
- Reduced bias by adding niche-specific full-length sequences to reference databases
- Accurate species detection via high phylogenetic resolution
- Cost-efficient, adaptable primer selection with reduced primer bias
Validated in diverse plant, soil, and marine sponge samples; benchmarked against metagenomics and commercial kits. CoSMIC has demonstrated superior resolution, reduced ambiguity, and high alignment accuracy in experimental settings.