The growing need for sustainable biofuel alternatives is challenged by the difficulty of breaking down lignin, a major component of plant biomass. Versatile Peroxidases (VPs) are enzymes naturally capable of degrading lignin, but their structural complexity makes them difficult to produce at scale. This technology presents a suite of engineered VPs with enhanced stability and activity that can be efficiently expressed in yeast, offering a practical and scalable solution for lignin processing and potentially an array of different compounds with applications across various industries.
Applications
- Biofuel production from lignocellulosic biomass
- Production of valuable aromatic compounds (e.g., vanillin) from lignin monomers for use in the food and fragrance industries
- Bioremediation of environmental pollutants and food detoxification
- Bleaching in the paper manufacturing and textile industry processing and textile industry processing, including oxidative dye treatments
- Diagnostic and biosensing tools using colorimetric peroxidase substrates (e.g., ABTS, DMP)
- Industrial biocatalysis under harsh pH, temperature, and oxidative conditions
- High expression yields in yeast
- Enhanced enzyme stability under industrial conditions
- Broad substrate specificity
- Enables the use of enzyme consortia for efficient lignin degradation
Four of 36 engineered VPs were successfully expressed in yeast, with three showing diverse activity, stability across pH and temperature conditions, and industrial potential.
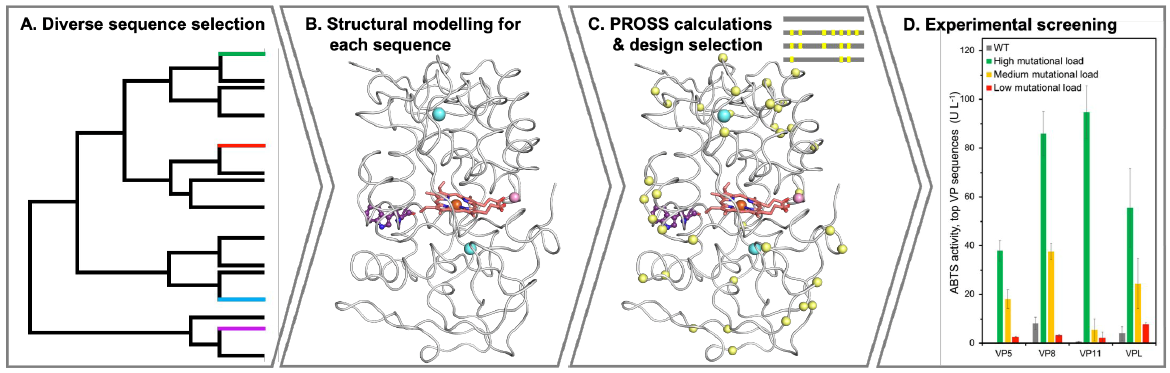
Workflow for computational enzyme optimization combining deep learning-based structure prediction, one-shot PROSS stability design, and experimental screening, resulting in recombinant VPs with high stability, activity, and efficient expression in yeast.
.Barber-Zucker S, Mindel V, Garcia-Ruiz E, Weinstein JJ, Alcalde M, Fleishman SJ. Stable and Functionally Diverse Versatile Peroxidases Designed Directly from Sequences. J Am Chem Soc. 2022;144(8):3564-3571. doi:10.1021/jacs.1c12433


