Understanding the complex interplay between the human host, gut microbiome, and diet is critical for diagnosing and treating diseases like IBD, celiac disease, and metabolic disorders. However, current approaches, such as metagenomics and food diaries, are limited in resolving active functions or exposures. IPHOMED (Integrated Proteogenomics of HOst-Microbiome-Diet) is a metagenome-informed metaproteomics pipeline that concurrently quantifies dietary, microbial, and host proteins from a single sample with species-level resolution, enabling high-accuracy functional diagnostics and mechanistic insights into gastrointestinal and systemic disorders.
- Functional profiling of the microbiome in health and disease
- Protein biomarker discovery for diagnosis and treatment monitoring (e.g., IBD, celiac)
- Non-invasive tracking of dietary intake and compliance
- Gut barrier assessment from blood/urine samples
- Stool-based analysis of digestion and absorption
- Identification of therapeutic targets and bioactive peptides
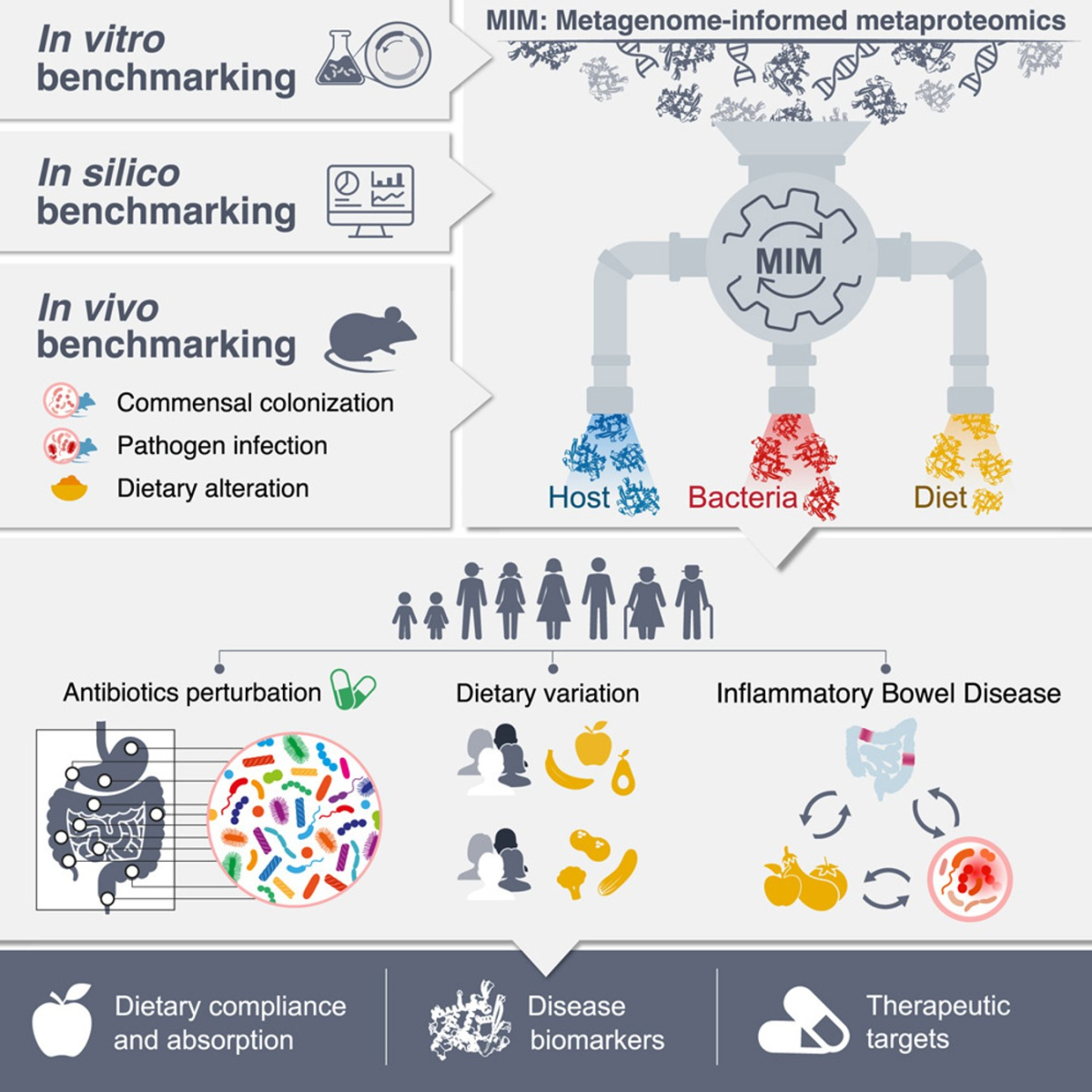
- Simultaneous quantification of host, microbial, and dietary proteins from the same sample
- Non-invasive and scalable; suits stool, blood, urine & mucosal samples
- Higher taxonomic resolution and accuracy than metagenomics or standard proteomics
- High-throughput discovery of diagnostic and therapeutic biomarkers
IPHOMED was validated in vitro, in vivo (mouse models), and in several human clinical cohorts.
It outperforms existing methods in diagnostic accuracy, with clinical-grade biomarker panels under development for IBD diagnosis, stratification, and dietary compliance monitoring.


